Abstract
Background/Purpose There is inherent difficulty with accurate diagnosis of bone marrow failure syndromes including Severe Aplastic Anemia (SAA), Paroxysmal Nocturnal Hemoglobinuria (PNH) and Hypoplastic Myelodysplastic Syndrome (MDS). We have attempted to identify and validate panels of disease-specific /disease-associated proteins biomarkers for their potential clinical value for accurate definition of patients presenting with features of bone marrow failure syndromes.
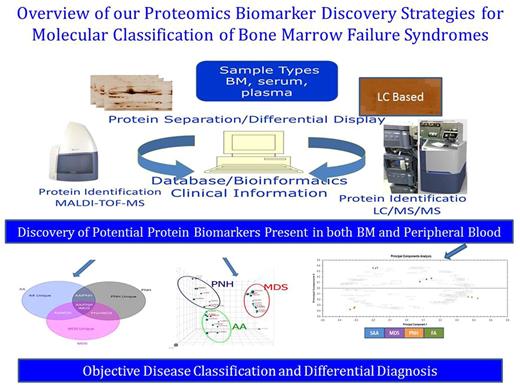
Methodology: Bone marrow plasma (MBP) and peripheral blood plasma (PBP) samples from 24 subjects with bone marrow failure; including AA/MDS/PNH/Fanconi Anemia (FA) as well as normal BMP and PBP were subjected to expression proteome analysis using label-free quantitative liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS).
Results: Over 900 unique protein species were identified of which approximately 124 and 174 were significantly differentially expressed (> 1.5- ∞- fold change & p < 0.05) in BMP and PBP respectively. These protein signatures independently discriminate patients into four distinct clusters; AA/MDS/PNH/FA using principal component analysis (PCA) (Figure 1). Approximately 25% of the identified proteins showed similar expression patterns across the two datasets from BMP and PBP. Ingenuity Pathway Analysis showed some of our identified proteins as potential biomarkers were implicated in hematological diseases and multiple signaling networks associated with known bone marrow failure syndrome genes.
In addition, six (6) of the identified proteins (THBS1, S100A9, GSTP1, GPX1, SOD2 and LMNB1) were independently confirmed as bone marrow failure associated genes using the 65 bone marrow failure syndromes gene panel from the curated CTD Gene-Disease Associations dataset (1). Interestingly, this differentially expressed protein panel were uniquely expressed across all disease sample groups as well as normal control sample group, thus making them potential biomarkers.
Conclusions: Our data indicates the usefulness of multivariate analysis of quantitative proteome data as a means of discovery of disease related or disease specific biomarkers for bone marrow syndromes. These proteins once validated, on a larger cohort of patients, might be valuable to complement the currently existing parameters for reliable and objective disease diagnosis, monitoring treatment response and clinical outcome of bone marrow failure syndrome patients.
Reference: 1. Andrew D. Rouillard Gregory W. Gundersen Nicolas F. Fernandez Zichen Wang Caroline D. Monteiro Michael G. McDermott Avi Ma'ayan, The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins ; Database (Oxford) (2016) 2016: baw100. DOI: https://sci-hub.cc/https://doi.org/10.1093/database/baw100
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal